
A Dictionary is a non-sequential data type that stores data in key-value pairs. This is in contrast to lists, where data is stored sequentially, and each value is associated with its location in the sequence. A key is a unique identifier for a piece of data, while a value is a piece of data associated with each key.
Examples of dictionaries include: * The English dictionary contains entries for unique words and their definition(s) - key: word, value: definition.
* The phonebook maps a person’s name (key) to their phone number (value) (or vice versa). * A English to Spanish translator would map english words (key) to their translation (value). * A post-office directory could map zip codes (key) to their city (value).
Why is this useful? For one, there are many examples of problems where naturally pair pieces of information (see the examples above). Second, with lists, we must use linear search in most circumstances to obtain information. If we have parallel lists, this can get tedious.
Say we want to store all user IDs and the associated user name for a class roster. We could use parallel lists (or a list of lists):
ids = ["asoni1","lfontes1",'lmeeden1']
names = ["Ameet Soni","Lila Fontes","Lisa Meeden"]To add a new user, we use append() and the item always goes at the end. We have to be careful to due the appends simultaneously so that the user’s information is at the same position in both lists:
ids.append("jknerr1")
names.append("Jeff Knerr")To find if a person is in our class, we could use linear search:
>>> "lfontes1" in ids
True
>>> "adanner1" in ids
FalseWhat if we want to get the user’s name given their id? This would require a linear search in ids to find the location, followed by indexing into names at the given location. Again, this is a linear search.
>>> loc = ids.index("lfontes1")
>>> names[loc]
'Lila Fontes'Note that Python makes this seem easy since it gave us methods for finding the location. This is still error prone - if we accidentally delete an item from only one list, the parallel lists are no longer useful.
Dictionaries can do this much more seamlessly. We will create a dictionary of key-value pairs where the key (unique identifier) is the user ID and the associated value is the user name.
users = {"asoni1":"Ameet Soni", "lfontes1":"Lila Fontes", "lmeeden1": "Lisa Meeden"}This initializes a dictionary (which uses curly braces as opposed to square brackets); each key-value pair is separated by a comma, and the key-values are designated with colon key:value.
To add a user, we simply index using the key and set its value.
users["jknerr1"] = "Jeff Knerr"That looks a lot like a list, except we don’t have to worry about an out-of-index error since it is non-sequential. Also, the index is not a location (integer), but the key itself.
To find if a user is in the class, we can use the same in operator to see if a key is in the dictionary (it cannot be used for values). While it looks the same, dictionaries are actually much faster since they don’t need linear search.
>>> "lfontes1" in users
True
>>> "adanner1" in users
FalseTo get a user’s name, we don’t need to find the index first. Dictionaries are built exactly for this type of query; they know where the name is by using the user id (key) as the identifier.
>>> users["lfontes1"]
'Lila Fontes'The central dogma of biology refers to the flow of genetic information from DNA to protein. A idea is the genetic code, which determines how DNA translates to protein (technically, it translates from RNA, but we’ll abstract that detail).

The building block of DNA is called a base, of which there are 4 (abbreviated A,C,G,T). The building block of proteins are amino acids, of which there are 20 types (also with one-letter abbreviations). Three consecutive bases of DNA - known as a codon - determines exactly one amino acid in the protein. So, our algorithm is to march down the DNA sequence, three characters at a time. Each three characters adds one amino acid to our sequence. Here is a table with the exact conversion rules:
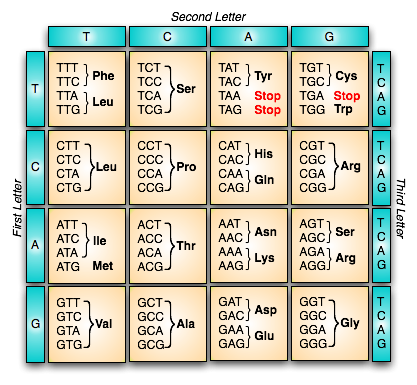
Note that there is a specific start codon (ATG) that is required to start the process; there are three stop codons that end the translation process. All other possible three-letter codons map to an amino acid.
In translate_DNA.py, I have provided a program to perform translation. The key idea here is to see how we use dictionaries to specify the mapping from codon (three DNA characters) to an amino acid. The program loads a DNA sequence file from disk (I have provided test.fasta and gfp.fasta which is the groundbreaking green fluorescent protein). The codon table is then loaded from disk (codon.txt) and a dictionary is created with codons being the key and the amino acid being the value. Lastly, the function translate() uses this codon table to determine how to translate from DNA to a protein.
Complete the readCodons function to add entries to the dictionary. Each line of the file has one amino acid and then all of the codons that map to it. Each codon/amino-acid pair will need to be in the dictionary.
Once that is complete, print our your dictionary to see if your codon table was loaded correctly (if there is time, format this output using the provided function)
Complete the inner-loop of translate, which will take the current codon and append its translation to the protein sequence. Be sure to check if you hit a stop codon (the stop codons are not in the dictionary); break the loop if you have.
Here is output for the test example:
$ python3 translate_dna.py
Enter filename: test.fasta
DNA sequence:
TTAATAGCGTGGAATGATCCTTATTAAAGAGTGTCACGAAGAGTCGGAATAGAATATGGAGGCGACAGTCGAGGGTGGGA
TAGAGTCCTAAAGATAACATTAAGTGTTAATCATTTTATTTTAAAA
Translated protein sequence:
ILIKECHEESESuppose we wanted to do some analysis on texts, such as Alice in Wonderland or Ulysses. We actually have these stored in files on our system including Alice in Wonderland (/usr/local/doc/alice.txt) and Mary Had a Little Lamb (/usr/local/doc/mary.txt).
The question we’ll ask is: how could we read these in and count the frequencies of all of the words?
I have provided a partially complete program, wordCount.py. Complete the following exercises:
Explain program with your neighbor. How do we process the file? What are the keys and values for our dictionary?
What is the role of updateFrequency?
In pairs complete updateFrequency to update the dictionary for each word read in.
Complete the print method such that all words in the dictionary with value greater than the value n are printed